Modeling Mycobacterial lipids and their action on host membranes
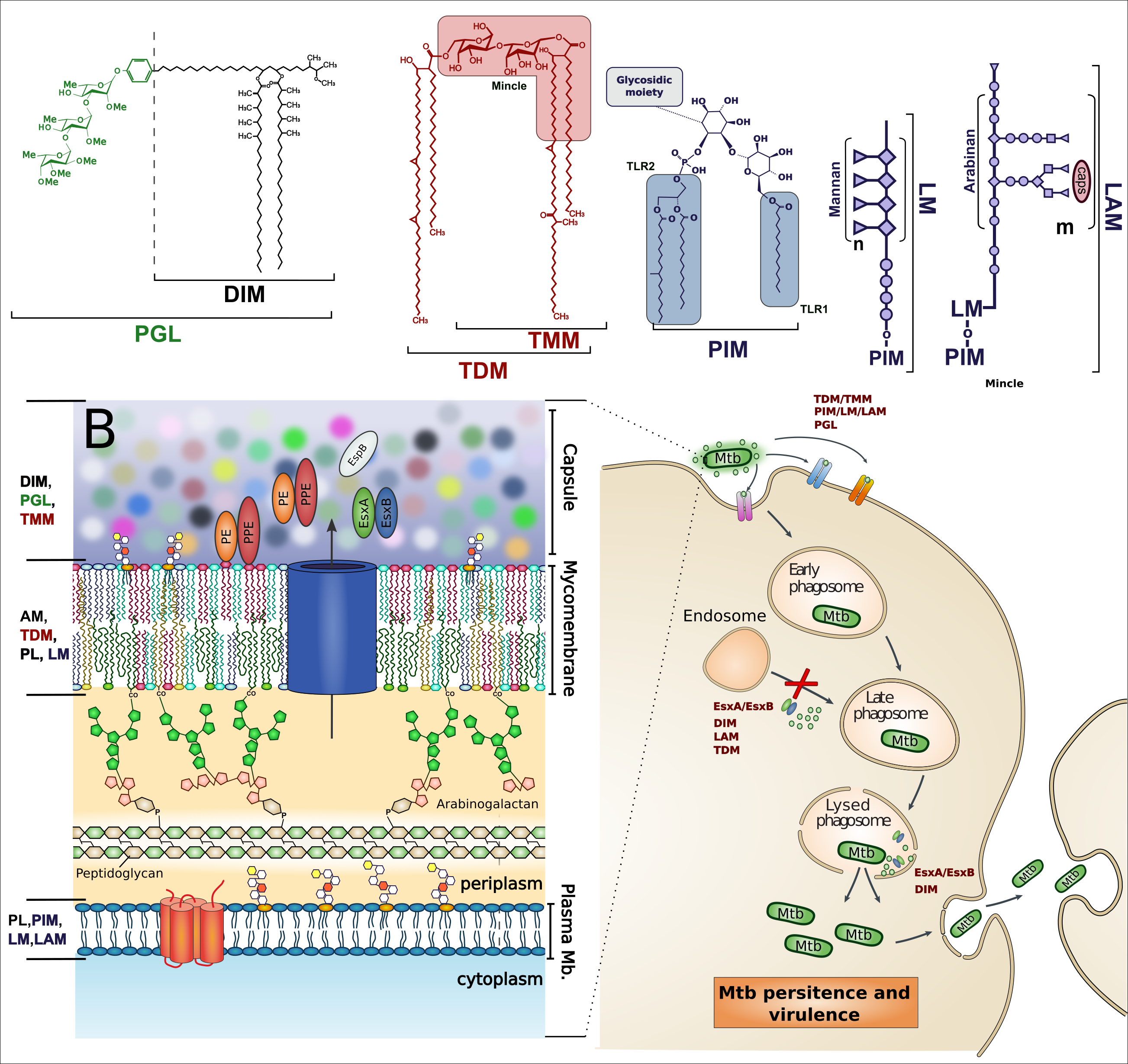
Mycobacterium tuberculosis (Mtb) is the main causative agent of the disease Tuberculosis (TB). These bacteria are considered as Gram-Positive bacteria that are distinguished by complex surface lipids. These lipids can be used as virulence effectors acting at the host membrane to damage it and modulate immune response. They are also the building blocks of a very thick and tight envelope which constitute a diffusion barrier to drugs, thus contributing to Mtb persistence. These lipids may also have a synergistic action with secreted mycobacterial proteins, which helps the pathogen to evade the host immune response. Thus, understanding the structure and functions of these complex molecules, and how they can act on or with different proteins, will help biologists to develop new drug strategies.
My aim is to develop models of Mtb lipids and their interactions with proteins to better understand how these molecules will act on the host membrane and modulate the immune response. This will be possible by developing a multi-scale strategy combining modeling with new experimental results.
Modeling supra-molecular membrane systems
Massively parallel molecular simulations can be used to understand the flexibility of huge macromolecular systems. Very long time-scale molecular simulations (on the order of milliseconds) are often required to accurately model several key biological phenomena. We have recently modelled very large membrane systems to see how clusters of Outer Membraner Proteins (OMPs) can form long moving frontiers [1].
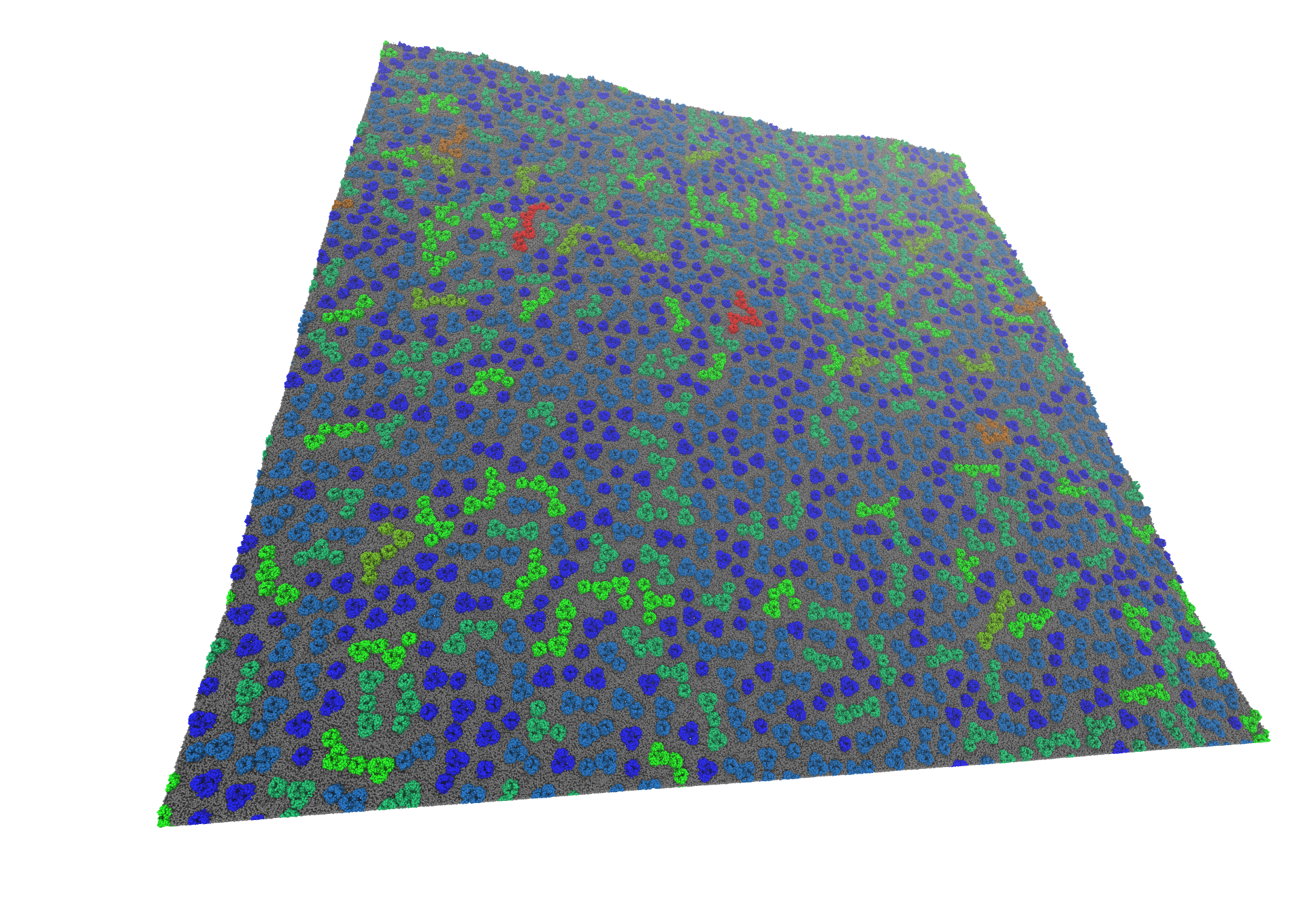
These very large systems necessitate the creation of new and efficient tools to analyse them. I am developing, in collaboration with computer scientists, different tools to display lipid flows inside the membrane [2] or interactions between membrane proteins and lipids.
